In a previous post, we looked at four different linear models for a collection of data where besides x and y values, the data was qualifies by an additional factor called a Group.
In those models, we either assumed that the difference in groups had no effect on the
model, models 1 and 2, or that the groups were independent, and we could estimate
their effects on slope and intercept individually. A compromise between these
two approaches is the mixed-effect model, sometimes called a multilevel linear
model or a random effects model. The difference is that the groups are assumed to
be a random sample from a larger population of groups measuring the same kinds of
data. In other words, are the groups we see all the groups we will see or is the data a sample from the world of possible groups?
A Linear Model
The
simplest mixed model is similar to model 2 from the previous post. In this model we expect the Group value to affect the intercept of the linear model.
The
difference from the similar linear model is the term ${\beta _{0j}} \sim N({\mu _{{\beta _0}}},\sigma _{{\beta _0}}^2)$. Here, rather than assuming ${\beta _0}$ is constant, we are assuming that the Group level indicators come from a
normal, but with unknown parameters, distribution. Mixed effects models use a maximum likelihood
method
to estimate the coefficients and the variance of the coefficient
In this
model, x is called a fixed effect and Group is called a random effect. However,
in statistical literature the terms have fluid meanings. (See https://stats.stackexchange.com/questions/4700/what-is-the-difference-between-fixed-effect-random-effect-and-mixed-effect-mode and https://statmodeling.stat.columbia.edu/2005/01/25/why_i_dont_use/ for
different definitions of fixed and random effects.) For our purposes, anything
inside the parenthesis in an R formula is a random effect. Group as a random
effect,
> library(lmerTest) > library(rsq) > lmm1 <- lmer(y ~ x + (1 | Group), data = df_test, REML = FALSE, control = lmerControl(optimizer = "Nelder_Mead")) > rsq(lmm1) # r-squared $model [1] 0.9431799 $fixed [1] 0.9201095 $random [1] 0.02307045 >
|
effect |
group |
term |
estimate |
std.error |
statistic |
df |
p.value |
Significance |
|
fixed |
NA |
(Intercept) |
0.425990318 |
0.049594947 |
8.589389522 |
32.91957856 |
6.44E-10 |
*** |
|
fixed |
NA |
x |
0.047049294 |
0.000430711 |
109.2364163 |
999.9529208 |
0 |
*** |
|
ran_pars |
Group |
sd__(Intercept) |
0.182703789 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Residual |
sd__Observation |
0.262164699 |
NA |
NA |
NA |
NA |
The table
shows estimates for the fixed effects, intercept and slope. For the random
effects we are provided with estimates of the standard deviation for Group and the
model, i.e. $\sigma _{{\beta _0}}^2$ and $\sigma _x^2$. The standard deviation of the intercept shows the
variability in the intercept across groups.
> write_mixed_model_coefs(lmm1, 'models/lmm1_coef.csv')
|
intercept |
x |
Group |
ranef_intercept |
|
0.365138253 |
0.047049294 |
1 |
-0.060852065 |
|
0.297419984 |
0.047049294 |
2 |
-0.128570334 |
|
0.430316754 |
0.047049294 |
3 |
0.004326436 |
|
0.429700924 |
0.047049294 |
4 |
0.003710606 |
|
0.659971258 |
0.047049294 |
5 |
0.23398094 |
|
0.793991226 |
0.047049294 |
6 |
0.368000908 |
|
0.395911819 |
0.047049294 |
7 |
-0.030078499 |
|
0.611620526 |
0.047049294 |
8 |
0.185630208 |
|
0.19373114 |
0.047049294 |
9 |
-0.232259178 |
|
0.353416766 |
0.047049294 |
10 |
-0.072573552 |
|
0.209006939 |
0.047049294 |
11 |
-0.216983379 |
|
0.641626397 |
0.047049294 |
12 |
0.215636079 |
|
0.487901608 |
0.047049294 |
13 |
0.06191129 |
|
0.091383548 |
0.047049294 |
14 |
-0.33460677 |
|
0.463582185 |
0.047049294 |
15 |
0.037591867 |
|
0.417185297 |
0.047049294 |
16 |
-0.008805021 |
|
0.256496571 |
0.047049294 |
17 |
-0.169493747 |
|
0.364360738 |
0.047049294 |
18 |
-0.06162958 |
|
0.40266712 |
0.047049294 |
19 |
-0.023323198 |
|
0.436768508 |
0.047049294 |
20 |
0.01077819 |
|
0.643599117 |
0.047049294 |
21 |
0.217608799 |
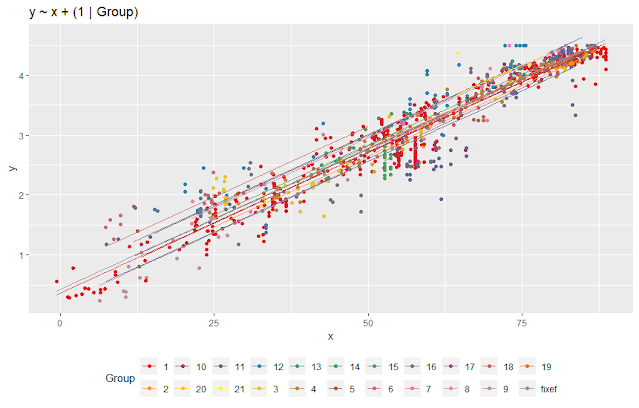
This plot appears
similar to the plot from lmm3 from the previos post., but the shift in intercept is slightly
less. The "fixef" line shows the fixed
effect intercept and slope.
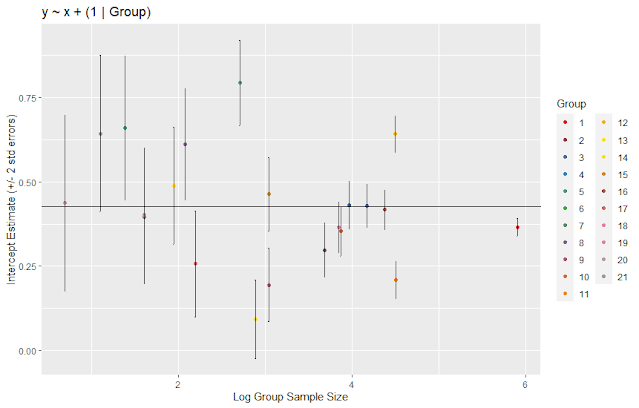
We can use
the estimates and the standard error of the random effects to examine the
effect of size. The plot shows the intercept estimates and +/- 2 standard
errors. The horizontal line is the fixed effect estimate of the intercept. From
the plot below, we see that labs with a small number of observations have
larger standard errors. Near log(size) = 4 (about 55 observations) the standard
errors are reduced, and the estimates are closer to the fixed effect estimate
of the intercept.
Varying Slope and Intercept
As with
the regression models, we can allow the slope to vary with Group. We will try this in two
different ways. We can model variation is the slope as independent of variation
in the intercept or as correlated with intercept as in a linear equation.
\[\begin{array}{l}{y_i} = {\beta _{0j}} + {\beta _{1j}}x + {\varepsilon _i}\\y \sim N({\beta _{0j}} + {\beta _{1j}}x,\sigma _x^2)\\{\beta _{0j}} \sim N({\mu _{{\beta _0}}},\sigma _{{\beta _0}}^2)\,\,{\beta _{1j}} \sim N({\mu _{{\beta _1}}},\sigma _{{\beta _1}}^2)\end{array}\]
In R,
> lmm2 <- lmer(y ~ x + (1 | Group) + (0 + x | Group), data = df_test, REML = FALSE, control = lmerControl(optimizer = "bobyqa")) > rsq(lmm2) $model [1] 0.9454617 $fixed [1] 0.9202339 $random [1] 0.02522783 >
|
effect |
group |
term |
estimate |
std.error |
statistic |
df |
p.value |
Significance |
|
fixed |
NA |
(Intercept) |
0.441912771 |
0.056084539 |
7.879404526 |
20.52537169 |
1.23E-07 |
*** |
|
fixed |
NA |
x |
0.046776353 |
0.000627214 |
74.5780174 |
6.799725224 |
3.68E-11 |
*** |
|
ran_pars |
Group |
sd__(Intercept) |
0.198195525 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Group.1 |
sd__x |
0.001170298 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Residual |
sd__Observation |
0.260799199 |
NA |
NA |
NA |
NA |
In this case, there are standard deviations for random effects for slope and intercept. We see that the random effect of Group on x covers only a very small proportion of the total variance while the effect of Group on the intercept covers more.
We can
plot the individual labs.
Finally,
we can model the effect of Group on both the slope and intercept.
> lmm3 <- lmer(y ~ x + (1 + x | Group), data = df_test, REML = TRUE, control = lmerControl(optimizer = "Nelder_Mead")) > rsq(lmm3) $model [1] 0.9472098 $fixed [1] 0.9208767 $random [1] 0.02633312 >
|
effect |
group |
term |
estimate |
std.error |
statistic |
df |
p.value |
Significance |
|
fixed |
NA |
(Intercept) |
0.436631488 |
0.082690437 |
5.280314164 |
16.73261814 |
6.45E-05 |
*** |
|
fixed |
NA |
x |
0.046688799 |
0.00078598 |
59.40201396 |
14.9205669 |
3.85E-19 |
*** |
|
ran_pars |
Group |
sd__(Intercept) |
0.334591939 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Group |
cor__(Intercept).x |
-1 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Group |
sd__x |
0.002807165 |
NA |
NA |
NA |
NA |
|
|
ran_pars |
Residual |
sd__Observation |
0.259399193 |
NA |
NA |
NA |
NA |
Again, the small standard deviation of the random effect on x indicates that this effect covers only a small proportion of the total variance (0.07%).
The plot shows
more variation than the previous one, but much less than the similar regression
model.
The plot below
shows the random effects for intercept and x along with +/- their standard
deviations.
Testing the Null Hypothesis
We have three nested mixed models:
lmm1: y ~ x + (1 | Group)lmm2: y ~ x + (1 | Group) + (0 + x |Group)lmm3: y ~ x + (1 + x| Group)
As before, we can perform use anova to test the hypothesis that simpler models are better.
> anova(lmm1, lmm2, lmm3) Data: df_test Models: lmm1: y ~ x + (1 | Group) lmm2: y ~ x + (1 | Group) + (0 + x | Group) lmm3: y ~ x + (1 + x | Group) npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) lmm1 4 220.04 239.67 -106.021 212.04 lmm2 5 220.94 245.48 -105.470 210.94 1.101 1 0.294 lmm3 6 200.45 229.90 -94.225 188.45 22.491 1 2.112e-06 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
We see
that we can reject the null hypothesis, at least for lmm3, given the small p-values. In addition,
AIC, BIC, and the loglikelihood for lmm3 show that the more complex model is an
improvement over the simpler models.
Code for these models can be found on GitHub.



No comments:
Post a Comment